

National Geographic Store. Promethease - SNPedia. The easiest way to make a report is to visit promethease.com This takes about 10 minutes and costs $5.
Video of how to make and read a Promethease report An example report based on either What is Promethease? [edit] Promethease is a literature retrieval system that builds a personal DNA report based on the scientific literature cited in SNPedia and a file of genotype (DNA) data. Read about the features of Promethease. Examples[edit] Math.mun. Available utilities for processing unzipped autosomal files from Family Tree DNA and/or 23andMe: These utilities were developed by David Pike.

My original motivation for developing these utilities was so that I could privately perform some advanced analysis of autosomal DNA results, with my objective being to better pursue genealogical research within my own family. Instead of limiting these utilities to my own personal use, I have made them available in the hope that they might assist other members of the genetic genealogy community with their own individual research goals. Given the do-it-yourself nature of the intended user, the utilities are presented with minimal documentation and without verbose explanation of analysis results. Also note that the analysis performed by these utilities is based on my own methodology and implementation, whereas Family Tree DNA, 23andMe and AncestryDNA have their own proprietary analysis methodologies.
Some other notes about these utilities: MtDNA Haplogroup Analysis. GEDmatch. Family Tree DNA - Genetic Genealogy Starts Here. Mitosearch. Ysearch. 23andMe - Genetic Testing for Health, Disease & Ancestry; DNA Test. NOVA: Cracking Your Genetic Code. DNA Testing by AncestryDNA reveals Ancestry and Genealogy. The Genographic Project by National Geographic - Human Migration, Population Genetics. Dodecad Ancestry Project. How to make your own calculator for DIYDodecad. As I have explained in the README file of DIYDodecad, it is possible to use the software to create and distribute new calculators, based on different marker sets/ancestral populations.
(The following discussion will only be useful to other genome bloggers, or people who have experience with ADMIXTURE software). Currently, DIYDodecad is distributed together with the 'dv3' calculator ("Dodecad v3"). This consists of a set of files: dv3.par (The parameter file that tells DIYDodecad what to expect and what to do) dv3.alleles (Allele names and variants) dv3.12.F (Allele frequencies for 12 ancestral populations) dv3.txt (Names for 12 ancestral populations) I will now explain how you can use PLINK and ADMIXTURE to create your own calculator. (1) Running ADMIXTURE In the following discussion, I will assume that you have your dataset in binary PLINK format (bed/bim/fam files), that it has 123,456 markers, and you run ADMIXTURE regularly for 7 populations, e.g .
CAVEAT! . (2) Preparing the test.alleles file. 'K12b' and 'K7b' calculators. I am releasing two new calculators with K=12 and K=7 components, named 'K12b' and 'K7b'.
You can scroll down to the bottom if you are just interested in the downloads, or read on.New Features. 'K10a' calculator. The 'K10a' calculator represents an intermediate stage between the K7 and K12 analyses released so far from the Project.
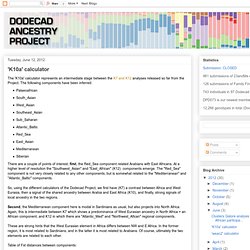
The following components have been inferred:Palaeoafrican South_Asian West_Asian Southeast_Asian Sub_Saharan Atlantic_BalticRed_Sea East_Asian Mediterranean Siberian There are a couple of points of interest; first, the Red_Sea component related Arabians with East Africans. At a higher level of resolution the "Southwest_Asian" and "East_African" (K12) components emerge. The "Red_Sea" component is not very closely related to any other components, but is somewhat related to the "Mediterranean" and "Atlantic_Baltic" components. So, using the different calculators of the Dodecad Project, we first have (K7) a contrast between Africa and West Eurasia, then a signal of the shared ancestry between Arabia and East Africa (K10), and finally, strong signals of local ancestry in the two regions.
Table of Fst distances between components: MDS plots of the first few dimensions: K35 calculator (no South Chinese cluster) K36 calculator. 'weac' calculator. This new calculator places individuals on the West Eurasian cline.
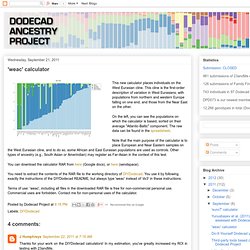
This cline is the first-order description of variation in West Eurasians, with populations from northern and western Europe falling on one end, and those from the Near East on the other. On the left, you can see the populations on which the calculator is based, sorted on their average "Atlantic-Baltic" component. The raw data can be found in the spreadsheet. Note that the main purpose of the calculator is to place European and Near Eastern samples on the West Eurasian cline, and to do so, some African and East Eurasian populations are used as controls. Other types of ancestry (e.g., South Asian or Amerindian) may register as Far-Asian in the context of this test. 'weac2' calculator. 'africa9' calculator. I have devised a new calculator targeted specifically for Africans.
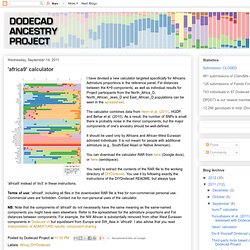
Admixture proportions in the reference panel, Fst distances between the K=9 components, as well as individual results for Project participants from the North_Africa_D, North_African_Jews_D and East_African_D populations can be seen in the spreadsheet. The calculator combines data from Henn et al. (2011), HGDP, and Behar et al. (2010).
As a result, the number of SNPs is small: there is probably noise in the minor components, but the major components of one's ancestry should be well-defined. It should be used only by Africans and African-West Eurasian admixed individuals. It is not meant for people with additional admixture (e.g., South/East Asian or Native American). 'world9' calculator. I have consistently received requests for an assessment of Amerindian ancestry.
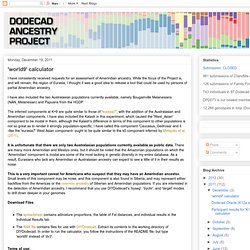
While the focus of the Project is, and will remain, the region of Eurasia, I thought it was a good idea to release a tool that could be used by persons of partial Amerindian ancestry. I have also included the two Australasian populations currently available, namely Bougainville Melanesians (NAN_Melanesian) and Papuans from the HGDP. The inferred components at K=9 are quite similar to those of 'eurasia7', with the addition of the Australasian and Amerindian components.
'euro7' calculator. I am releasing a new calculator for Europeans, including their immediate neighboring populations around the Black Sea (Caucasus and Anatolia).
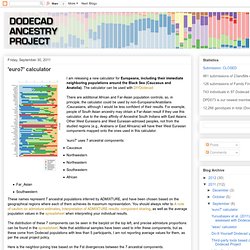
The calculator can be used with DIYDodecad There are additional African and Far-Asian population controls, so, in principle, the calculator could be used by non-Europeans/Anatolians/Caucasians, although I would be less confident of their results. For example, people of South Asian ancestry may obtain a Far-Asian result if they use this calculator, due to the deep affinity of Ancestral South Indians with East Asians.
Other West Eurasians and West Eurasian-admixed peoples, not from the studied regions (e.g., Arabians or East Africans) will have their West Eurasian components mapped onto the ones used in this calculator. 'globe4' calculator. Patterson et al. (2012) recently published evidence for admixture in northern Europeans between a population resembling modern Sardinians (and the Neolithic Tyrolean Iceman, whose genome was published earlier this year), and, surprisingly Native Americans.
The authors attribute the Amerindian-like ancestry element to a North Eurasian population that spawned Native Americans, and which also contributed ancestry to northern Europeans. They propose two possibilities for the origin of this admixture: (i) the Mesolithic Europeans resembled Amerindians, or (ii) there was an influx of Amerindian-like populations from the east during late prehistory. A palimpsest of these two processes may explain parts of the observed signal of admixture. In a recent K=4 admixture experiment, I demonstrated that ADMIXTURE software produces an Amerindian ancestral component that closely tracks the signal of admixture using the D-statistic test. 'globe10' calculator.
'globe13' calculator. 'bat' calculator (Balkans-Anatolia-Turkic) I have decided to make a new calculator for DIYDodecad that may be useful for individuals from the Balkans and Anatolia. You can download it from here at Google Docs (or here from sendspace). The terms of use are the same as for DIYDodecad v 2.0. To run it, you simply extract the contents of the RAR file in your working directory, and type bat.par whenever you typed dv3.par in the instructions. The reference populations can be seen below. I have included all available Balkan populations, as well as Turks and Armenians. The marker set is the same as used in Dodecad v3. The components have been named accordingly, but please note that they do not necessarily reflect recent ancestors. Likewise, the Turkic component is only part of the genetic makeup of the Turkic speakers who arrived in Anatolia, since those probably also carried West Eurasian population elements picked up en route from Siberia to Anatolia.
'eurasia7' calculator. This calculator was made with 196 different populations and 2,659 individuals, including 518 project participants. The following Dodecad populations do not have 5 individuals yet, so they are included in the OTHERS_D generic category: Algerian_D, North_African_Jews_D, Slovenian_D, Mixed_Scandinavian_D, Danish_D, Moroccan_D, Tunisian_D, Serb_D, Austrian_D, Saudi_D, Pakistani_D, Tatar_Various_D, Palestinian_D, Greek_Italian_D, Romanian_D, Swiss_German_D, Szekler_D, Mandaean_D, Azeri_D, Czech_D, Georgian_D, Belgian_D, Latvian_D, Estonian_D, Bangladesh_D, Yemenese_D, Sri_Lanka_D, Hungarian_D, Basque_D, Udmurt_D, Egyptian_D As always, I encourage people with 4 grandparents from the same country or ethnic group of Eurasia, North or East Africa to contact me (do not send data!) For possible inclusion in the Project. Do-It-Yourself Dodecad v 2.1.
Geno 2.0 patch for DIYDodecad. (See important update at the end of this post) People who have tested using the Genographic Project's Geno 2.0 test can now use the DIYDodecad tool with their data. The raw data download from this test has a slightly different format than the ones from 23andMe and Family Finder, so it is necessary to convert your data in a format that DIYDodecad can interpret. So, after you have downloaded and extracted the DIYDodecad software as per its instructions, you should also download a couple of extra files into your working directory; these files are included in this patch: standardize.r which replaces the standardize.r in the DIYDodecad software bundle, and allows you to convert your Geno 2.0 formatted datahgdp.base.txt which includes additional information about SNP markers that is not found in your Geno 2.0 raw data download, and which is necessary to complete the conversion process.
Standardize('johndoe.csv', company='geno2') where johndoe.csv is your unzipped raw data download.